Selected Publications
Here are some of my selected publications that I'm particularly proud of.
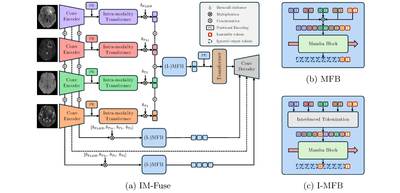
MICCAI - 2025
IM-Fuse: A Mamba-based Fusion Block for Brain Tumor Segmentation with Incomplete Modalities
Vittorio Pipoli, Alessia Saporita, Kevin Marchesini, Costantino Grana, Elisa Ficarra, Federico Bolelli
28th International Conference on Medical Image Computing and Computer Assisted Intervention, May 2025
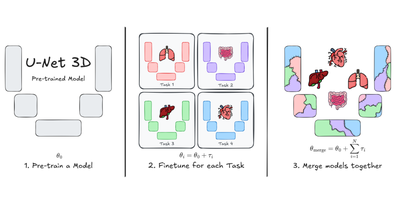
MICCAI - 2025
U-Net Transplant: The Role of Pre-training for Model Merging in 3D Medical Segmentation
Luca Lumetti, Giacomo Capitani, Elisa Ficarra, Costantino Grana, Simone Calderara, Angelo Porrello, Federico Bolelli
28th International Conference on Medical Image Computing and Computer Assisted Intervention, May 2025
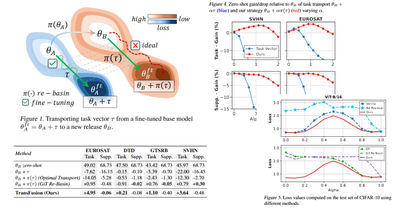
ICML - 2025
Update Your Transformer to the Latest Release: Re-Basin of Task Vectors
Filippo Rinaldi, Giacomo Capitani, Lorenzo Bonicelli, Angelo Porrello, Donato Crisostomi, Federico Bolelli, Emanuele Rodolà, Elisa Ficarra, Simone Calderara
Proceedings of the International Conference on Machine Learning, May 2025
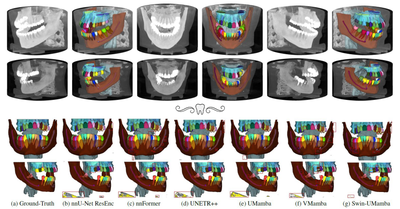
CVPR - 2025
Segmenting Maxillofacial Structures in CBCT Volumes
Federico Bolelli, Kevin Marchesini, Niels van Nistelrooij, Luca Lumetti, Vittorio Pipoli, Elisa Ficarra, Shankeeth Vinayahalingam, Costantino Grana
IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Mar 2025
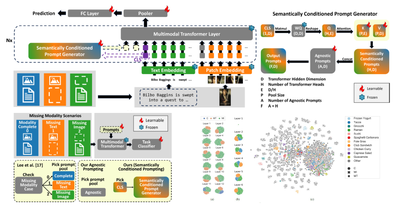
WACV - 2025
Semantically Conditioned Prompts for Visual Recognition under Missing Modality Scenarios
Vittorio Pipoli, Federico Bolelli, Sara Sarto, Marcella Cornia, Lorenzo Baraldi, Costantino Grana, Rita Cucchiara, Elisa Ficarra
IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) 2025
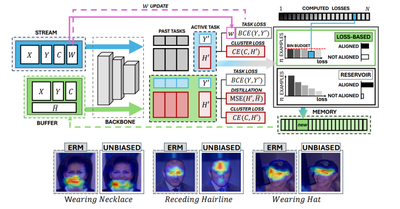
WACV - 2025
Towards Unbiased Continual Learning: Avoiding Forgetting in the Presence of Spurious Correlations
Giacomo Capitani, Lorenzo Bonicelli, Angelo Porrello, Federico Bolelli, Simone Calderara, Elisa Ficarra
IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) 2025

IEEE TMI - 2024
Segmenting the Inferior Alveolar Canal in CBCTs Volumes: the ToothFairy Challenge
Federico Bolelli, Luca Lumetti, Shankeeth Vinayahalingam, ..., Alexandre Anesi, Costantino Grana
IEEE Transactions on Medical Imaging, Dec 2024
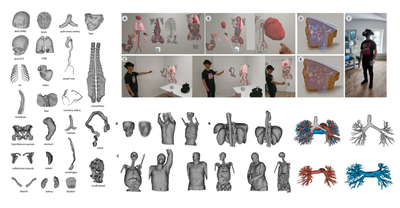
BMT - 2024
MedShapeNet – a large-scale dataset of 3D medical shapes for computer vision
Jianning Li, ..., Federico Bolelli, Costantino Grana, Luca Lumetti, ..., Jan Egger
Biomedical Engineering/Biomedizinische Technik, Sep 2024
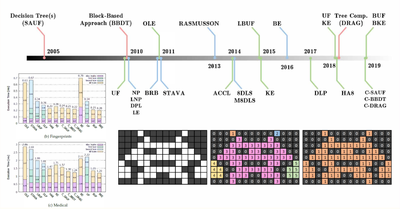
IEEE TPDS - 2024
A State-of-the-Art Review with Code about Connected Components Labeling on GPUs
Federico Bolelli, Stefano Allegretti, Luca Lumetti, Costantino Grana
IEEE Transactions on Parallel and Distributed Systems 2024
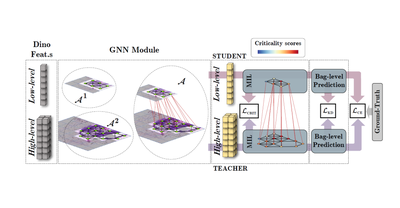
MICCAI - 2023
DAS-MIL: Distilling Across Scales for MIL Classification of Histological WSIs
Gianpaolo Bontempo, Angelo Porrello, Federico Bolelli, Simone Calderara, Elisa Ficarra
Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, Oct 2023
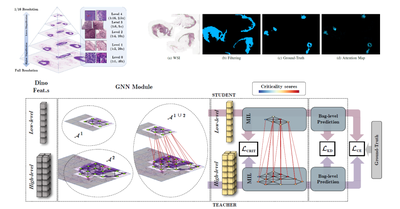
TMI - 2023
A Graph-Based Multi-Scale Approach with Knowledge Distillation for WSI Classification
Gianpaolo Bontempo, Federico Bolelli, Angelo Porrello, Simone Calderara, Elisa Ficarra
IEEE Transactions on Medical Imaging 2023
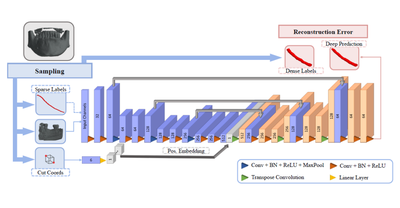
CVPR - 2022
Improving Segmentation of the Inferior Alveolar Nerve through Deep Label Propagation
Marco Cipriano, Stefano Allegretti, Federico Bolelli, Federico Pollastri, Costantino Grana
IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Jun 2022
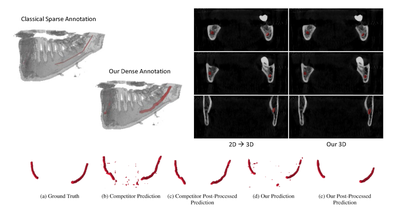
IEEEAccess - 2022
Deep Segmentation of the Mandibular Canal: a New 3D Annotated Dataset of CBCT Volumes
Marco Cipriano, Stefano Allegretti, Federico Bolelli, ..., Alexandre Anesi, Costantino Grana
IEEE Access 2022
TPAMI - 2021
One DAG to Rule Them All
Federico Bolelli, Stefano Allegretti, Costantino Grana
IEEE Transactions on Pattern Analysis and Machine Intelligence, Jan 2021
TIP - 2019
Spaghetti Labeling: Directed Acyclic Graphs for Block-Based Connected Components Labeling
Federico Bolelli, Stefano Allegretti, Lorenzo Baraldi, Costantino Grana
IEEE Transactions on Image Processing, October 2019
CAIP - 2019
How does Connected Components Labeling with Decision Trees perform on GPUs?
Stefano Allegretti, Federico Bolelli, Michele Cancilla, Federico Pollastri, Laura Canalini, Costantino Grana
Computer Analysis of Images and Patterns, Sep 2019
TPDS - 2019
Optimized Block-Based Algorithms to Label Connected Components on GPUs
Stefano Allegretti, Federico Bolelli, Costantino Grana
IEEE Transactions on Parallel and Distributed Systems, August 2019
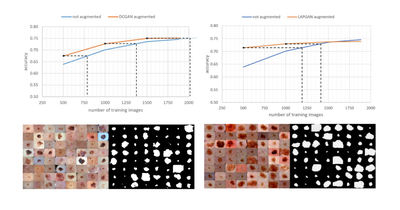
MTAP - 2019
Augmenting Data with GANs to Segment Melanoma Skin Lesions
Federico Pollastri, Federico Bolelli, Roberto Paredes, Costantino Grana
Multimedia Tools and Applications, May 2019
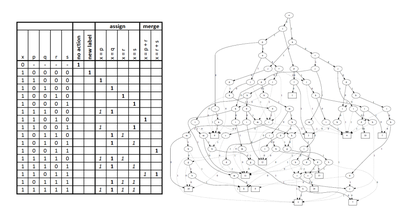
ICPR - 2018
Connected Components Labeling on DRAGs
Federico Bolelli, Lorenzo Baraldi, Michele Cancilla, Costantino Grana
2018 24th International Conference on Pattern Recognition (ICPR), Aug 2018
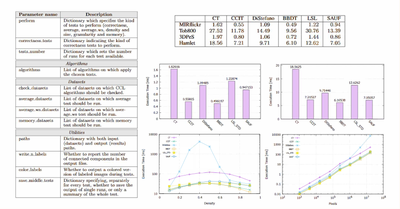
ICPR - 2016
YACCLAB - Yet Another Connected Components Labeling Benchmark
Costantino Grana, Federico Bolelli, Lorenzo Baraldi, Roberto Vezzani
2016 23rd International Conference on Pattern Recognition (ICPR), Dec 2016
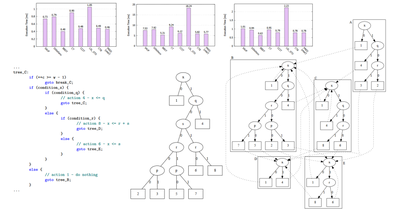
ACIVS - 2016
Optimized Connected Components Labeling with Pixel Prediction
Costantino Grana, Lorenzo Baraldi, Federico Bolelli
Advanced Concepts for Intelligent Vision Systems, Oct 2016